Ralstonia solanacearum
| Ralstonia solanacearum | |
|---|---|
| |
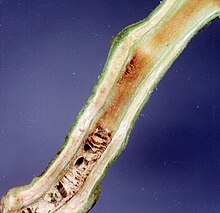
| Damage caused by Ralstonia solanacearum on tomato stem | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Pseudomonadota |
| Class: | Betaproteobacteria |
| Order: | Burkholderiales |
| Family: | Burkholderiaceae |
| Genus: | Ralstonia |
| Species: | R. solanacearum
|
| Binomial name | |
| Ralstonia solanacearum | |
| Type strain | |
| ATCC 11696 CCUG 14272 | |
| Synonyms | |
|
Burkholderia solanacearum (Smith 1896) Yabuuchi et al. 1993 | |
Ralstonia solanacearum is an aerobic non-spore-forming, Gram-negative, plant pathogenic bacterium. R. solanacearum is soil-borne and motile with a polar flagellar tuft. It colonises the xylem, causing bacterial wilt in a very wide range of potential host plants. It is known as Granville wilt when it occurs in tobacco. Bacterial wilts of tomato, pepper, eggplant, and Irish potato caused by R. solanacearum were among the first diseases that Erwin Frink Smith proved to be caused by a bacterial pathogen. Because of its devastating lethality, R. solanacearum is now one of the more intensively studied phytopathogenic bacteria, and bacterial wilt of tomato is a model system for investigating mechanisms of pathogenesis.[2][3] Ralstonia was until recently classified as Pseudomonas,[1] with similarity in most aspects,[4] except that it does not produce fluorescent pigment like Pseudomonas.[5] The genomes from different strains vary from 5.5 Mb up to 6 Mb, roughly being 3.5 Mb of a chromosome and 2 Mb of a megaplasmid.[6] While the strain GMI1000 was one of the first phytopathogenic bacteria to have its genome completed,[7] the strain UY031 was the first R. solanacearum to have its methylome reported.[6] Within the R. solanacearum species complex, the four major monophyletic clusters of strains are termed phylotypes, that are geographically distinct: phylotypes I-IV are found in Asia, the Americas, Africa, and Oceania, respectively.[2][6]
Ralstonia solanacearum was once considered as a possible biological control agent for Kahili ginger (Hedychium gardnerianum), a highly invasive species.[8] However, R. solanacearum is no longer used as a biological control for Kahili ginger in Hawaiian forests because of its wide host range. The ginger-parasitizing strain will infect numerous ginger species, including edible ginger (Zingiber officinale), shampoo ginger (Z. zerumbet), and pink and red ginger (Alpinia purpurata).[9]
Hosts and symptoms
[edit]
Hosts
[edit]Plant hosts that R. solanacearum infects include:
- Crops
- Potato (Solanum tuberosum); tomato (Lycopersicum esculentum);[10] soybean (Glycine max);[11] eggplant/aubergine (Solanum melongena); banana, (Musa spp); geranium (Pelargonium spp.); ginger (Zingiber officinale); tobacco (Nicotiana tabacum); bell pepper/sweet pepper (Capsicum spp.); olive (Olea europea); rose (Rosa).[citation needed]
- Wild hosts
- Woody nightshade (Solanum dulcamara)
Symptoms
[edit]Geranium:[12]
- Wilting begins with lower leaves and petioles and works its way up the plant.
- Wilted leaves have chlorotic, wedge-shaped areas or chlorotic and/or necrotic leaf margins. No leaf spots are evident.
- Eventually, the entire plant collapses on the medium.
- White runny material oozes from cut stems.
Potato:[12]
- Wilting of the leaves occurs at the end of the day with recovery at night. Plants eventually fail to recover and die.
- Brown staining of vascular ring happens and pus may exude from the ring when the tuber is squeezed.
- Pale ooze may exude from eyes and heel end of potato. Soil will adhere to the oozing eyes.
- Cross-section of a stem placed in water will exude milky white strands.
- Unlike the fungal wilts, the leaves remains green in bacterial wilt.
Disease cycle
[edit]Survival
[edit]Ralstonia solanacearum can overwinter in plant debris or diseased plants, wild hosts, seeds, or vegetative propagative organs (other germplasm) like tubers. The bacteria can survive for a long time in water (up to 40 years at 20–25 °C (68–77 °F) in pure water), and the bacterial population is reduced in extreme conditions (temperature, pH, salts, e.g.). Infected land sometimes cannot be used again for susceptible crops for several years. R. solanacearum can also survive in cool weather and enter a state of being viable but not culturable. In most cases, this stage is not an agricultural threat because the bacteria usually become avirulent after recovering.[2]
Dispersal
[edit]Ralstonia solanacearum causes wilting at high populations (108 – 1010 cfu/g tissue) and disperses in several routes. The large number of R. solanacearum can shed from roots of symptomatic and nonsymptomatic plants. Besides that, bacterial ooze (which is usually used as a sign for detection) on plant surfaces) can enter the surrounding soil or water, contaminating farming equipment or may be acquired by insect vectors.[2] In addition, this pathogen can be spread by contaminated flood water, irrigation, contaminated tools, or infected seeds. In northern Europe, the pathogen has become established in solanaceous weeds which grow in slow-moving rivers. When such contaminated water is used to irrigate potatoes, the pathogen enters the potato production system. The race 3 biovar 2 strain can survive in perennial nightshades which act as secondary hosts, and can also cause bacterial wilt of tomato.[13] Some EU states and Middle Eastern countries have not yet been able to eradicate this pathogen.[citation needed]
Infection
[edit]Ralstonia solanacearum usually enters the plant by a wound. Natural wounds (created by abscission of flowers, genesis of lateral roots) and unnatural ones (by agricultural practices or nematodes and xylem-feeding insects) could become entry sites for R. solanacearum. The bacteria get access to the wounds partially by flagellar-mediated swimming motility and chemotaxic attraction toward root exudates. Unlike many phytopathogenic bacteria, R. solanacearum potentially requires only one entry site to establish a systemic infection that results in bacterial wilt.[2]
After invading a susceptible host, R. solanacearum multiplies and moves systemically within the plant before bacterial wilt symptoms occur. Wilting should be considered as the most visible side effect that usually occurs after extensive colonization of the pathogen. When the pathogen gets into the xylem through natural openings or wounds, tyloses may form to block the axial migration of bacteria within the plant. In susceptible plants, this sometimes happens slowly and infrequently to prevent pathogen migration, and may instead lead to vascular dysfunction by unspecifically obstructing uncolonized vessels.[clarification needed]
Wilting occurs at high bacterial populations in the xylem and is partially due to vascular dysfunction in which sufficient water cannot reach the leaves. At this time, extracellular polysaccharide (EPS1) content is about 10 μg/g tissue in the taproot, hypocotyl, and midstem; EPS1 concentration is higher later on at more than 100 μg/g tissue in fully wilted plant. Ralstonia's systemic toxin also causes loss of stomatal control, but no evidence shows excessive transpiration as its consequence. The primary factor contributing to wilting is probably blocking of pit membranes in the petioles and leaves by the high molecular weight EPS1. High bacterial densities are byproducts of plant cell wall degradation; tyloses and gums produced by the plant itself are other contributing factors to wilting.[2]
Natural genetic transformation
[edit]Most strains of R. solanacearum are competent for genetic transformation.[14] Natural genetic transformation is a sexual process involving DNA transfer from one bacterial cell to another through the intervening medium, and the integration of the donor sequence into the recipient genome by homologous recombination. R. solanacearum is able to exchange large DNA fragments ranging from 30 to 90 thousand bases.[14]
Virulence mechanisms
[edit]Ralstonia solanacearum possesses genes for all six protein secretion pathways that have been characterized in Gram-negative bacteria. Perhaps the best-studied of these is the Type III secretion system (T3SS or TTSS), which secretes infection-promoting effector proteins (T3Es) into host cells. Around 74 suspected or confirmed T3Es have been identified in R. solanacearum to date, although the functions of very few are currently known. Despite being just one of several protein secretion systems, T3SS is necessary for R. solanacearum to cause disease.[15] No single effector protein has been found to significantly alter pathogenicity of R. solanacearum, but simultaneous disruption of certain subsets of effectors (such as the set of seven GALA effectors in strain GMI1000) strongly affects virulence of the pathogen. For example, GALA 7 is necessary for virulence on Medicago truncatula, hinting that T3E diversity may play a role in determining the broad host range of the R. solanacearum species complex.[16]
The type III secretion system is not unique to R. solanacearum, and is, in fact, very ancient. The evolutionary history of the T3SS is contested; a high degree of similarity to the flagellum has sparked debate over the relationship between these two structures.[2]
About half of T3SS proteins are highly conserved in R. solanacearum and likely constitute a very old and stabilized group of effectors in the core genome of the species complex.[17]
Environment
[edit]The environment in which R. solanacearum is commonly found is affected by the particular race (a genetically diverse population within a species), and the particular biovar (a strain that differs physiologically or biochemically from other strains.) Race 1, race 2 biovar 1, and race 3, biovar 2 are three of the most common and important strains. Race 1 strains have a broad host range including tobacco and bananas, and are usually found in tropical and subtropical environments, as they have trouble surviving cooler temperatures, and are endemic to the southeastern United States.[20] Race 2 strains have a more limited host range than race 1, and are mostly restricted to tropical environments. Race 3 strains are more cold tolerant than the other two and are found in tropical highlands and temperate areas.[20] The host range for race 3 biovar 2 includes potatoes, tomatoes, and geraniums. Race 3 biovar 2 is very common throughout the world, but is not generally reported in North America,[21] so is the focus of many sanitation and quarantine management practices to prevent the introduction or spread of the pathogen.
Although it is not there yet, researchers at the University of Guam are concerned about the possible spread of R. solanacearum to Guam.[22]
Management
[edit]General management
[edit]Commercial chemicals have generally proven to be ineffective in controlling the pathogen and are not recommended as a means of control.[2] In regions where the pathogen is established, a strategy of integrated disease management is the best method to reduce any impact of the pathogen. Using pathogen-free planting materials is a necessity. Planting resistant cultivars minimizes the ill effects of the pathogen, although no completely immune cultivars are now available. Finally, a good rotation system that follows susceptible crops with resistant or nonhost crops can assist in diminishing the pathogen.[2] The pathogen is listed as a select agent in the United States; if the pathogen is detected by a proper authority, a number of management protocols may be implemented. These can range from surveys to quarantines of infected and potentially infected plant material, which in turn may lead to larger eradication and sanitation programs.[20]
Specific host plant symptoms and management
[edit]Potato
[edit]Wilting and yellowing of the leaves, as well as overall stunting of the plant, are typical symptoms.[23] The leaves may also take on a bronze cast[24] along with stems becoming streaked and tuber eyes becoming discolored. Tubers also start to rot if left in the ground. A milky-white sticky exudate or ooze, consisting of bacterial cells and their extracellular polysaccharide, is usually noticeable in freshly cut-sections of infected tubers.[25] Control of R. solanacearum is difficult because it is a soil borne pathogen, has wide host range, long survival in the soil, and has wide biological variation. No single control method has been found to be 100% effective, although in locations where the pathogen is established, some level of bacterial wilt control has been possible through use of a combination of diverse methods.[25] These methods include phytosanitation and cultural practices, chemical control, biological control, and host resistance. General sanitation practices are recommended to prevent spread of the disease, as chemical control is ineffective. Crop rotation with resistant crops is useful, as is altering the pH of the soil to keep it low in the summer (4-5), and higher in the fall (6.)[24]
Tomato
[edit]Younger leaves of the plant will become flaccid, and adventitious roots may appear on the stem of the plant. The vascular system exhibits a progressively darker brown color as the disease progresses, in addition to possible lesions on the stem.[26] Management practices are similar to those of potato.[citation needed]
Banana
[edit]
Commonly known as Moko disease, after a banana variety from Trinidad that went extinct in the 1890s. Typically, yellowing and wilting of older leaves occurs, as well as reduced fruit size and eventual rotting of the fruit.[27] In addition, flowers can become blackened and shriveled, and the vascular tissue discolored.[28] Exclusion of the disease where it is not present is the only effective means of control. If an area does become infected, all of the infected plants must be eliminated, which is why strong sanitation practices must be used to reduce the spread of disease.[28]
Importance
[edit]Ralstonia solanacearum is classified as one of the world's most important phytopathogenic bacteria due to its lethality, persistence, wide host range, and broad geographic distribution. Although the pathogen causes major yield losses in the tropics and subtropics, it is currently a continuing threat in temperate climates.[2]
Ralstonia solanacearum is a high-profile alien plant pathogen of A2 quarantine status affecting a very wide range of crops. This means that it is present in parts of Europe, but is under statutory control. Worldwide, the most important crops affected are: potato, tomato, tobacco, banana, and geranium. In the UK and the rest of the EU, the most important crops affected are potato and tomato. It would cause serious economic damage were it to become more established than it currently is. Losses are due to actual yield reduction and also due to statutory measures taken to eliminate the disease.[citation needed]
Bacterial wilt caused by R. solanacearum is of economic importance because it infects over 250 plant species in over 50 families. As of 2007, this pathogen has affected over 450 host species representing 54 plant families due to its broad host range around the world.[29] The disease is known as southern wilt, bacterial wilt, and brown rot of potato. Many more dicots suffer from the disease than do monocots. Among the monocot hosts, the order Zingiberales dominates, with five of nine families being infected by this bacterium.[2] The reason why some families are more susceptible to bacterial wilt is still unknown. Originally, R. solanacearum is found in tropical, subtropical, and warm, temperate climates, but is not believed to survive cold temperatures. However, this pathogen has recently been detected in geraniums (Pelargonium spp.) in Wisconsin, USA [30] and was traced back to the import of geranium cuttings to North America and Europe from the highland tropics where race 3 biovar 2 is endemic [31]
Brown rot of potato caused by R. solanacearum race 3 biovar 2 is among the most serious disease of potato worldwide, and is responsible for an estimated $950 million in losses each year.[32] Race 3 biovar 2 is cold tolerant and classified as a quarantine pathogen.[31] In addition, this race/biovar has been listed as a select agent in the Agricultural Bioterrorism Act of 2002, and is considered to have potential to be developed as a bioterror weapon.[30]
See also
[edit]References
[edit]- ^ a b Yabuuchi, Eiko; Kosako, Yoshimasa; Yano, Ikuya; Hotta, Hisako; Nishiuchi, Yukiko (1995-11-01). "Transfer of Two Burkholderia and An Alcaligenes Species to Ralstonia Gen. Nov". Microbiology and Immunology. 39 (11): 897–904. doi:10.1111/j.1348-0421.1995.tb03275.x. ISSN 1348-0421. PMID 8657018.
- ^ a b c d e f g h i j k Denny T., "Plant Pathogenic Ralstonia species" in GNANAMANICKAM, S. S. (2006). Plant-associated bacteria. Dordrecht, Springer. pp 1-62
- ^ Vailleau, Fabienne; Genin, Stéphane (2023-09-05). "Ralstonia solanacearum : An Arsenal of Virulence Strategies and Prospects for Resistance". Annual Review of Phytopathology. 61 (1): 25–47. doi:10.1146/annurev-phyto-021622-104551. ISSN 0066-4286. PMID 37506349.
- ^ Agrios, George (2005). Plant pathology. Burlington, MA: Elsevier Academic Press. pp. 647–649. ISBN 978-0-08-047378-9. OCLC 134821046.
- ^ Agrios, George N. (2005). Plant pathology (5th ed.). Amsterdam: Elsevier Academic Press. p. 622. ISBN 0-12-044565-4. OCLC 55488155.
- ^ a b c Guarischi-Sousa, Rodrigo; Puigvert, Marina; Coll, Núria S.; Siri, María Inés; Pianzzola, María Julia; Valls, Marc; Setubal, João C. (15 January 2016). "Complete genome sequence of the potato pathogen Ralstonia solanacearum UY031". Standards in Genomic Sciences. 11 (1): 7. doi:10.1186/s40793-016-0131-4. PMC 4714475. PMID 26779304.
- ^ Salanoubat, M.; Genin, S.; Artiguenave, F.; Gouzy, J.; Mangenot, S.; Arlat, M.; Billault, A.; Brottier, P.; Camus, J. C.; Cattolico, L.; Chandler, M.; Choisne, N.; Claudel-Renard, C.; Cunnac, S.; Demange, N.; Gaspin, C.; Lavie, M.; Moisan, A.; Robert, C.; Saurin, W.; Schiex, T.; Siguier, P.; Thébault, P.; Whalen, M.; Wincker, P.; Levy, M.; Weissenbach, J.; Boucher, C. A. (31 January 2002). "Genome sequence of the plant pathogen Ralstonia solanacearum". Nature. 415 (6871): 497–502. doi:10.1038/415497a. PMID 11823852.
- ^ Andersona, R. C.; Gardner, D. E. (1999). "An Evaluation of the Wilt-Causing Bacterium Ralstonia solanacearum as a Potential Biological Control Agent for the Alien Kahili Ginger (Hedychium gardnerianum) in Hawaiian Forests". Biological Control. 15 (2): 89–96. doi:10.1006/bcon.1999.0705.
- ^ Paret, M.L.; de Silva, A.S.; Criley, R.A.; Alvarez, A.M. (2008). "Ralstonia solanacearum Race 4:Risk Assessment for Edible Ginger". HortTechnology. 18: 90–96. doi:10.21273/HORTTECH.18.1.90.
- ^ Farahat, M.G.; Abdel Rahman, Tahany, M.; Hussein, R.A.; Zaghlol, Gihan M. (2016). "Biological Control of Tomato Bacterial Wilt Disease by Endophytic Pseudomonas fluorescens and Bacillus subtilis". Egyptian Journal of Botany. 56 (2): 543–558. doi:10.21608/ejbo.2017.1150. ISSN 2357-0350.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Terblanche, J.; de Villiers, D.A. (2013). Prior, Philippe; Allen, Caitilyn; Elphinstone, John (eds.). Bacterial Wilt Disease: Molecular and Ecological Aspects (1st ed.). Paris: Springer Science & Business Media. p. 326. ISBN 9783662035924.
- ^ a b "Bacterial Wilt- Ralstonia solanacearum race 3 biovar 2". Massnrc.org. 2008-02-25. Retrieved 2012-09-24.
- ^ Charkowski, Amy (2020). "Bacterial Diseases of Potato". The Potato Crop. pp. 351–388. doi:10.1007/978-3-030-28683-5_10. ISBN 978-3-030-28682-8.
- ^ a b Coupat B, Chaumeille-Dole F, Fall S, Prior P, Simonet P, Nesme X, Bertolla F (2008). "Natural transformation in the Ralstonia solanacearum species complex: number and size of DNA that can be transferred". FEMS Microbiol. Ecol. 66 (1): 14–24. doi:10.1111/j.1574-6941.2008.00552.x. PMID 18662313.
- ^ Vasse, J; Genin, S.; Pascal, F.; Boucher, C.; Brito, B. (2000). "The hrpB and hrpG Regulatory Genes of Ralstonia solanacearum Are Required for Different Stages of the Tomato Root Infection Process". Molecular Plant-Microbe Interactions. 13 (3): 259–267. doi:10.1094/mpmi.2000.13.3.259. PMID 10707351.
- ^ Angot, Aurelie; Peeters, N; Lechner, E; Vailleu, F; Baud, C; Gentzbittel, L; Sartorel, E; Genschik, P; Boucher, C; Genin, Stephane (2006). "Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants". Proceedings of the National Academy of Sciences. 103 (39): 14620–14625. Bibcode:2006PNAS..10314620A. doi:10.1073/pnas.0509393103. PMC 1600009. PMID 16983093.
- ^ Remenant, Benoit; Coupat-Goutaland, B.; Guidot, A.; Cellier, G.; Prior, P. (2010). "Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence". BMC Genomics. 11 (1): 379. doi:10.1186/1471-2164-11-379. PMC 2900269. PMID 20550686.
- ^ Genin, S.; Boucher, C. (2004). "Lessons learned from the genome analysis of Ralstonia solanacearum". Annual Review of Phytopathology. 42: 107–134. doi:10.1146/annurev.phyto.42.011204.104301. PMID 15283662.
- ^ Heuer, H.; Yin, Y. -N.; Xue, Q. -Y.; Smalla, K.; Guo, J. -H. (2007). "Repeat Domain Diversity of avrBs3-Like Genes in Ralstonia solanacearum Strains and Association with Host Preferences in the Field". Applied and Environmental Microbiology. 73 (13): 4379–4384. Bibcode:2007ApEnM..73.4379H. doi:10.1128/AEM.00367-07. PMC 1932761. PMID 17468277.
- ^ a b c "R. solanacearum/Bacterial wilt - Brown rot of potato". Plantpath.ifas.ufl.edu. 2008-09-12. Retrieved 2012-09-24.
- ^ "Archived copy" (PDF). Archived from the original (PDF) on 2010-10-07. Retrieved 2010-12-08.
{{cite web}}: CS1 maint: archived copy as title (link) - ^ "UOG watches for moth, bacteria that attack nightshade plants". 12 October 2020.
- ^ "Potato brown rot symptoms". Cals.ncsu.edu. Retrieved 2012-09-24.
- ^ a b "Archived copy" (PDF). Archived from the original (PDF) on 2010-10-09. Retrieved 2010-12-08.
{{cite web}}: CS1 maint: archived copy as title (link) - ^ a b Champoisea et al. 2009 (2009). "Ralstonia solanacearum Race 3 Biovar 2 Causes Tropical Losses and Temperate Anxieties". Plant Health Progress. 10: 35. doi:10.1094/PHP-2009-0313-01-RV. S2CID 13013168.
{{cite journal}}: CS1 maint: numeric names: authors list (link) - ^ "Tomato bacterial wilt symptoms". Cals.ncsu.edu. Retrieved 2012-09-24.
- ^ "Banana Moko disease symptoms". Cals.ncsu.edu. Retrieved 2012-09-24.
- ^ a b "Archived copy" (PDF). Archived from the original (PDF) on 2012-03-14. Retrieved 2010-12-08.
{{cite web}}: CS1 maint: archived copy as title (link) - ^ Wicker, Emmanuel (2007-08-24). "Ralstonia Solanacearum Strains from Martinique (French West Indies) Exhibiting a New Pathogenic Potential". Applied and Environmental Microbiology. 73 (21): 6790–6801. Bibcode:2007ApEnM..73.6790W. doi:10.1128/AEM.00841-07. PMC 2074947. PMID 17720825.
- ^ a b Hudelson B. 2005. University of Wisconsin Pest Alert - Ralstonia wilt.
- ^ a b Milling A., Meng F., Denny T., Allen C. 2009. Interactions with hosts at cool temperatures, not cold tolerance, explain the unique epidemiology of Ralstonia solanacearum race 3 biovar 2
- ^ Ephinstone, J. G. 2005. The current bacterial wilt situation: a global overview. pp 9-28 in: Bacterial Wilt: The Disease and the Ralstonia solanacearum Species Complex. C. Allen, P. Prior, and A. C. Hayward, eds. American Phytopathological Society, St. Paul, MN.
- Poueymiro, M.; Genin, S. (2009). "Secreted proteins from Ralstonia solanacearum: a hundred tricks to kill a plant". Current Opinion in Microbiology. 12 (1): 44–52. doi:10.1016/j.mib.2008.11.008. PMID 19144559.
- ^ p. 45, "Nearly half of the GMI1000 effector repertoire (34/74) appears to be conserved within the species, and thus are deemed ancient or stably inherited along with the core genome (see Table 2)."
- ^
p. 49, "Type III effectors that trigger plant resistance
In response to pathogen attack, plants have evolved specific resistance ‘guard’ proteins able to recognize the presence or the action of some T3Es historically called ‘avirulence factors’. This specific recognition leads to the T3E-triggered immunity which mainly results in the HR (reviewed in [40]). To date, three R. solanacearum T3Es recognized as ‘avirulence’ proteins were characterized: the avrA gene product triggers HR elicitation on several tobacco species [41,42], while resistance to GMI1000 in specific Petunia and Arabidopsis ecotypes is due to the recognition of the PopP1 [20] or PopP2 proteins [43], respectively."
External links
[edit]- Species Profile - Southern Bacterial Wilt (Ralstonia solanacearum), National Invasive Species Information Center, United States National Agricultural Library. Lists general information and resources for Southern Bacterial Wilt.
- List of Plant Pathogens subject to Export Control
- Type strain of Ralstonia solanacearum at BacDive - the Bacterial Diversity Metadatabase
- Moorman, Gary W. (April 2011). "Bacterial Wilt - Ralston solanacearum". PennState Extension.
