Alphavirus
This article has multiple issues. Please help improve it or discuss these issues on the talk page. (Learn how and when to remove these messages)
|
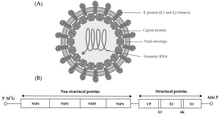
Alphavirus is a genus of RNA viruses, the sole genus in the Togaviridae family. Alphaviruses belong to group IV of the Baltimore classification of viruses, with a positive-sense, single-stranded RNA genome.[1] There are 32 alphavirus species, which infect various vertebrates such as humans, rodents, fish, birds, and larger mammals such as horses, as well as invertebrates.[1] Alphaviruses that can infect both vertebrates and arthropods are referred dual-host alphaviruses, while insect-specific alphaviruses such as Eilat virus and Yada yada virus are restricted to their competent arthropod vector.[2] Transmission between species and their vertebrate hosts (including human) occurs mainly via mosquitoes, making the alphaviruses a member of the collection of arboviruses – or arthropod-borne viruses.[1] Alphavirus particles are enveloped, have a 70 nm diameter, tend to be spherical (although slightly pleomorphic), and have a 40 nm isometric nucleocapsid.[3]
Genome
[edit]This section needs additional citations for verification. (May 2023) |
| Alpha_E1_glycop | |||||||||
|---|---|---|---|---|---|---|---|---|---|
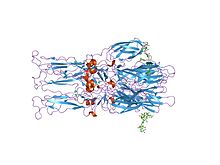 Crystal structure of the homotrimer of fusion glycoprotein E1 from Semliki Forest virus | |||||||||
| Identifiers | |||||||||
| Symbol | Alpha_E1_glycop | ||||||||
| Pfam | PF01589 | ||||||||
| InterPro | IPR002548 | ||||||||
| SCOP2 | 1rer / SCOPe / SUPFAM | ||||||||
| TCDB | 1.G | ||||||||
| OPM superfamily | 109 | ||||||||
| OPM protein | 1rer | ||||||||
| |||||||||
| Alpha_E2_glycop | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Mapping the E2 glycoprotein of alphaviruses | |||||||||
| Identifiers | |||||||||
| Symbol | Alpha_E2_glycop | ||||||||
| Pfam | PF00943 | ||||||||
| InterPro | IPR000936 | ||||||||
| TCDB | 1.G | ||||||||
| OPM superfamily | 109 | ||||||||
| OPM protein | 2yew | ||||||||
| |||||||||
| Alpha_E3_glycop | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Alpha_E3_glycop | ||||||||
| Pfam | PF01563 | ||||||||
| InterPro | IPR002533 | ||||||||
| TCDB | 1.G | ||||||||
| OPM superfamily | 109 | ||||||||
| |||||||||
The alphaviruses are small, spherical, enveloped viruses with a genome of a single strand of positive-sense RNA. The total genome length ranges between 11,000 and 12,000 nucleotides, and has a 5’ cap and a 3’ poly-A tail. The four non-structural protein genes are encoded in the 5′ two-thirds of the genome, while the three structural proteins are translated from a subgenomic mRNA colinear with the 3′ one-third of the genome.
There are two open reading frames (ORFs) in the genome, nonstructural and structural. The first is non-structural and encodes proteins (nsP1–nsP4) necessary for transcription and replication of viral RNA. The second encodes three structural proteins: the core nucleocapsid protein C, and the envelope proteins P62 and E1, which associate as a heterodimer. The viral membrane-anchored surface glycoproteins are responsible for receptor recognition and entry into target cells through membrane fusion.
Structural proteins
[edit]The proteolytic maturation of P62 into E2 and E3 causes a change in the viral surface. Together the E1, E2, and sometimes E3, glycoprotein "spikes" form an E1/E2 dimer or an E1/E2/E3 trimer, where E2 extends from the centre to the vertices, E1 fills the space between the vertices, and E3, if present, is at the distal end of the spike.[4] Upon exposure of the virus to the acidity of the endosome, E1 dissociates from E2 to form an E1 homotrimer, which is necessary for the fusion step to drive the cellular and viral membranes together. The alphaviral glycoprotein E1 is a class II viral fusion protein, which is structurally different from the class I fusion proteins found in influenza virus and HIV. The structure of the Semliki Forest virus revealed a structure that is similar to that of flaviviral glycoprotein E, with three structural domains in the same primary sequence arrangement.[5] The E2 glycoprotein functions to interact with the nucleocapsid through its cytoplasmic domain, while its ectodomain is responsible for binding a cellular receptor. Most alphaviruses lose the peripheral protein E3, but in Semliki viruses it remains associated with the viral surface.
Nonstructural proteins
[edit]Four nonstructural proteins (nsP1–4) which are produced as a single polyprotein constitute the virus' replication machinery.[6] The processing of the polyprotein occurs in a highly regulated manner, with cleavage at the P2/3 junction influencing RNA template use during genome replication. This site is located at the base of a narrow cleft and is not readily accessible. Before cleavage, nsP3 creates a ring structure that encircles nsP2. These two proteins have an extensive interface.
Mutations in nsP2 that produce noncytopathic viruses or a temperature sensitive phenotypes cluster at the P2/P3 interface region. P3 mutations opposite the location of the nsP2 noncytopathic mutations prevent efficient cleavage of P2/3. This in turn affects RNA infectivity altering viral RNA production levels.
Virology
[edit]The virus has a 60–70 nanometer diameter. It is enveloped, spherical and has a positive-strand RNA genome of ~12 kilobases. The genome encodes two polyproteins. The first polyprotein consists of four non-structural units: in order from the N terminal to the C terminal - nsP1, nsP2, nsP3, and nsP4. The second is a structural polyprotein composed of five expression units: from the N terminal to the C terminal - Capsid, E3, E2, 6K and E1. A sub genomic positive strand RNA - the 26S RNA - is replicated from a negative-stranded RNA intermediate. This serves as template for the synthesis of viral structural proteins. Most alphaviruses have conserved domains involved in regulation of viral RNA synthesis.
The nucleocapsid, 40 nanometers in diameter, contains 240 copies of the capsid protein and has a T = 4 icosahedral symmetry. The E1 and E2 viral glycoproteins are embedded in the lipid bilayer. Single E1 and E2 molecules associate to form heterodimers. The E1–E2 heterodimers form one-to-one contacts between the E2 protein and the nucleocapsid monomers. The E1 and E2 proteins mediate contact between the virus and the host cell.
Several receptors have been identified. These include prohibitin, phosphatidylserine, glycosaminoglycans and ATP synthase β subunit (ref needed).
Replication occurs within the cytoplasm, specifically in areas termed "spherules" separated by plasma membrane invaginations from the rest. Each complex occupies one such area of about 50-nm in inner diameter.[7]
Virions mature by budding through the plasma membrane, where virus-encoded surface glycoproteins E2 and E1 are assimilated. These two glycoproteins are the targets of numerous serologic reactions and tests including neutralization and hemagglutination inhibition. The alphaviruses show various degrees of antigenic cross-reactivity in these reactions and this forms the basis for the seven antigenic complexes, 32 species and many subtypes and varieties. The E2 protein is the site of most neutralizing epitopes, while the E1 protein contains more conserved, cross-reactive epitopes.
Evolution
[edit]A study of this taxon suggests that this group of viruses had a marine origin—specifically the Southern Ocean—and that they have subsequently spread to both the Old and New World.[8]
There are three subgroups in this genus: the Semliki Forest virus subgroup (Semliki Forest, O'nyong-nyong and Ross River viruses); the eastern equine encephalitis virus subgroup (eastern equine encephalitis and Venezuelan equine encephalitis viruses) and the Sindbis virus subgroup.[9] Sindbis virus, geographically restricted to the Old World, is more closely related to the eastern equine encephalitis subgroup, which are New World viruses, than it is to the Semliki Forest virus subgroup which is also found in the Old World.
Taxonomy
[edit]The following species are assigned to the genus:[10]
- Aura virus
- Barmah Forest virus
- Bebaru virus
- Caaingua virus
- Cabassou virus
- Chikungunya virus
- Eastern equine encephalitis virus
- Eilat virus
- Everglades virus
- Fort Morgan virus
- Getah virus
- Highlands J virus
- Madariaga virus
- Mayaro virus
- Middelburg virus
- Mosso das Pedras virus
- Mucambo virus
- Ndumu virus
- O'nyong'nyong virus
- Pixuna virus
- Rio Negro virus
- Ross River virus
- Salmon pancreas disease virus
- Semliki Forest virus
- Sindbis virus
- Southern elephant seal virus
- Tonate virus
- Trocara virus
- Una virus
- Venezuelan equine encephalitis virus
- Western equine encephalitis virus
- Whataroa virus
The seven complexes are:
- Barmah Forest virus complex
- Eastern equine encephalitis complex
- Eastern equine encephalitis virus (seven antigenic types)
- Middelburg virus complex
- Ndumu virus complex
- Semliki Forest virus complex
- Bebaru virus
- Chikungunya virus
- Getah virus
- Mayaro virus
- Subtype: Una virus
- O'nyong'nyong virus
- Subtype: Igbo-Ora virus
- Ross River virus
- Subtype: Sagiyama virus
- Semliki Forest virus
- Subtype: Me Tri virus
- Venezuelan equine encephalitis complex
- Cabassou virus
- Everglades virus
- Mosso das Pedras virus
- Mucambo virus
- Paramana virus
- Pixuna virus
- Rio Negro virus
- Trocara virus
- Subtype: Bijou Bridge virus
- Venezuelan equine encephalitis virus
- Western equine encephalitis complex
- Recombinants within this complex
- Buggy Creek virus
- Fort Morgan virus
- Highlands J virus
- Western equine encephalitis virus
- Unclassified
- Eilat virus
- Mwinilunga alphavirus
- Salmonid Alphavirus
- Southern elephant seal virus
- Tonate virus
- Caaingua virus[11]
Notes
[edit]Barmah Forest virus is related to the Semliki Forest virus. Middelburg virus, although classified as a separate complex, may be a member of the Semliki Forest virus group.
It seems likely that the genus evolved in the Old World from an insect-borne plant virus.[12]
Sindbis virus may have originated in South America.[13] The equine encephalitis viruses and the Sindbis virus are related.
The Old World and New World viruses appears to have diverged between 2000 and 3000 years ago.[14] Divergence between the Venezuelan equine encephalitis virus and the eastern equine virus appears to have been ~1400 years ago.[15]
The fish infecting clade appears to be basal to the other species.
The southern elephant seal virus appears to be related to the Sinbis clade.
Pathogenesis and immune response
[edit]| Virus | Human Disease | Vertebrate Reservoir | Distribution |
|---|---|---|---|
| Barmah Forest virus |
|
Humans | Australia |
| Chikungunya virus | Rash, arthritis | Primates, humans | Africa, Latin America, India, SE Asia |
| Eastern equine encephalitis virus | Encephalitis | Birds | Americas |
| Mayaro virus | Rash, arthritis | Primates, humans | South America |
| O'nyong'nyong virus | Rash, arthritis | Primates, Humans | Africa |
| Ross River virus | Rash, arthritis | Mammals, humans | Australia, South Pacific |
| Semliki Forest virus | Rash, arthritis | Birds | Africa |
| Sindbis virus | Rash, arthritis | Birds | Europe, Africa, Australia |
| Tonate virus | Encephalitis | Humans | South America |
| Una virus | Rash, arthritis | Primates, humans | South America |
| Venezuelan equine encephalitis virus | Encephalitis | Rodents, horses | Americas |
| Western equine encephalitis virus | Encephalitis | Birds, mammals | North America |
There are many alphaviruses distributed around the world with the ability to cause human disease. Infectious arthritis, encephalitis, rashes and fever are the most commonly observed symptoms. Larger mammals such as humans and horses are usually dead-end hosts or play a minor role in viral transmission; however, in the case of Venezuelan equine encephalitis the virus is mainly amplified in horses. In most other cases the virus is maintained in nature in mosquitoes, rodents and birds.
Terrestrial alphavirus infections are spread by insect vectors such as mosquitoes. Once a human is bitten by the infected mosquito, the virus can gain entry into the bloodstream, causing viremia. The alphavirus can also get into the CNS where it is able to grow and multiply within the neurones. This can lead to encephalitis, which can be fatal.
When an individual is infected with this particular virus, its immune system can play a role in clearing away the virus particles. Alphaviruses are able to cause the production of interferons. Antibodies and T cells are also involved. The neutralizing antibodies also play an important role to prevent further infection and spread.
Diagnosis, prevention, and control
[edit]Diagnoses is based on clinical samples from which the virus can be easily isolated and identified. There are no alphavirus vaccines currently available. Vector control with repellents, protective clothing, breeding site destruction, and spraying are the preventive measures of choice.[citation needed]
Use of alphaviruses for gene therapy and vaccination
[edit]Alphaviruses are of interest to gene therapy researchers, in particular the Ross River virus, Sindbis virus, Semliki Forest virus, and Venezuelan equine encephalitis virus have all been used to develop viral vectors for gene delivery. Of particular interest are the chimeric viruses that may be formed with alphaviral envelopes and retroviral capsids. Such chimeras are termed pseudotyped viruses. Alphaviral envelope pseudotypes of retroviruses or lentiviruses are able to integrate the genes that they carry into the expansive range of potential host cells that are recognized and infected by the alphaviral envelope proteins E2 and E1. The stable integration of viral genes is mediated by the retroviral interiors of these vectors.
There are limitations to the use of alphaviruses in the field of gene therapy due to their lack of targeting, however, through the introduction of variable antibody domains in a non-conserved loop in the structure of E2, specific populations of cells have been targeted. Furthermore, the use of whole alphaviruses for gene therapy is of limited efficacy both because several internal alphaviral proteins are involved in the induction of apoptosis upon infection and also because the alphaviral capsid mediates only the transient introduction of mRNA into host cells. Neither of these limitations extend to alphaviral envelope pseudotypes of retroviruses or lentiviruses. However, the expression of Sindbis virus envelopes may lead to apoptosis, and their introduction into host cells upon infection by Sindbis virus envelope pseudotyped retroviruses may also lead to cell death. The toxicity of Sindbis viral envelopes may be the cause of the very low production titers realized from packaging cells constructed to produce Sindbis pseudotypes.
Another branch of research involving alphaviruses is in vaccination. Alphaviruses are can be engineered to create replicon vectors which efficiently induce humoral and T-cell immune responses. In these contexts, replicons have been termed self-amplifying RNA.[16] The replicons lack the ability to transmit to new cells as they lack the sequences encoding for structural proteins, which are replaced with a gene of interest such as an antigen for generation of immunity against a disease. They could therefore be used to vaccinate against viral, bacterial, protozoan, and tumor antigens.
History
[edit]Initially, the Togaviridae family included what are now called the Flaviviruses, within the Alphavirus genus. The flaviviruses were formed into their own family when sufficient differences with the alphaviruses were noted due to the development of sequencing.[17] Rubella virus was formerly included in the family Togaviridae in its own genus Rubivirus, but is now classified in its own family Matonaviridae.[18] Alphavirus is now the sole genus in the family.
- 1930 – Western equine encephalitis virus is first isolated in the United States (the first alphavirus ever isolated)
- 1933 – Eastern equine encephalitis virus is first isolated in the United States.
- 1938 – Venezuelan equine encephalitis is isolated.
- 1941 – Western equine encephalitis epidemic is seen in the United States. It affects 300,000 horses and 3,336 humans.
- 1941 – Norman Gregg notices large number of children with cataracts following a rubella outbreak. This and other defects are then categorized under the congenital rubella syndrome.
- 1942 – Semliki Forest virus is isolated in Buliyama, Bwamba County, Uganda.
- 1952 – Sindbis virus is isolated in the Sindbis health district, 40 mi (64 km) north of Cairo, Egypt.
- 1959 – Ross River virus is isolated from Aedes vigilax mosquitoes (now known as Ochlerotatus vigilax)[19] which were trapped at the Ross River in Australia.
- 1963 – Ross River virus, which causes epidemic polyarthritis (mostly seen in Australia), is isolated by Doherty and colleagues.[20]
- 1971 – Last epidemic of Venezuelan equine encephalitis is seen in horses in southern Texas.[21]
- 1986 – Barmah Forest virus is identified as causing human disease in Australia.[22]
- 2001 – Scientists solved the crystal structure of the glycoprotein shell of the Semliki Forest virus.
- 2005–2006 – Large epidemic of the chikungunya virus on the island of La Réunion and the surrounding islands in the Indian Ocean[23]
- 2006 – Major epidemic of the chikungunya virus in India with over 1.5 million cases reported[24]
See also
[edit]Sources
[edit]- "Arboviruses". Virology-online.
- "ICTV Sources". ICTV. Archived from the original on 12 February 2006.
- Smerdou, C.; Liljestrom, P. (2000). "Alphavirus vectors: from protein production to gene therapy". Gene Therapy and Regulation. 1 (1): 33–63. doi:10.1163/156855800744520. ISSN 1568-5586.
- Rayner JO, Dryga SA, Kamrud KI (2002). "Alphavirus vectors and vaccination". Reviews in Medical Virology. 12 (5): 279–96. doi:10.1002/rmv.360. PMID 12211042. S2CID 21432844.
- Rhême, Céline; Ehrengruber, Markus U.; Grandgirard, Denis (2005). "Alphaviral cytotoxicity and its implication in vector development". Experimental Physiology. 90 (1): 45–52. doi:10.1113/expphysiol.2004.028142. PMID 15542620.
- Schmaljohn, Alan L.; McClain, David (1996). "54. Alphaviruses (Togaviridae) and Flaviviruses (Flaviviridae)". In Baron, Samuel (ed.). Medical Microbiology (4th ed.). University of Texas Medical Branch at Galveston. ISBN 0-9631172-1-1. PMID 21413253. NBK7633.
References
[edit]- ^ a b c Ahola, Tero; McInerney, Gerald; Merits, Andres (1 January 2021), Kielian, Margaret; Mettenleiter, Thomas C.; Roossinck, Marilyn J. (eds.), "Chapter Four - Alphavirus RNA replication in vertebrate cells", Advances in Virus Research, vol. 111, Academic Press, pp. 111–156, doi:10.1016/bs.aivir.2021.07.003, retrieved 20 August 2024
- ^ Elrefaey AM, Abdelnabi R, Rosales Rosas AL, Wang L, Basu S, Delang L (31 August 2020). "Understanding the Mechanisms Underlying Host Restriction of Insect-Specific Viruses". Viruses. 12 (9): 964. doi:10.3390/v12090964. PMC 7552076. PMID 32878245.
- ^ Chen R, Mukhopadhyay S, Merits A, Bolling B, Nasar F, Coffey LL, et al. (June 2018). "ICTV Virus Taxonomy Profile: Togaviridae". The Journal of General Virology. 99 (6): 761–2. doi:10.1099/jgv.0.001072. PMID 29745869.
- ^ Vénien-Bryan C, Fuller SD (February 1994). "The organization of the spike complex of Semliki Forest virus". J. Mol. Biol. 236 (2): 572–83. doi:10.1006/jmbi.1994.1166. PMID 8107141.
- ^ Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, Wengler G, Rey FA (April 2001). "The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH". Cell. 105 (1): 137–48. doi:10.1016/S0092-8674(01)00303-8. PMID 11301009. S2CID 16535677.
- ^ Shin G, Yost SA, Miller MT, Elrod EJ, Grakoui A, Marcotrigiano J (2012). "Structural and functional insights into alphavirus polyprotein processing and pathogenesis". Proc Natl Acad Sci U S A. 109 (41): 16534–9. Bibcode:2012PNAS..10916534S. doi:10.1073/pnas.1210418109. PMC 3478664. PMID 23010928.
- ^ Spuul, P; Balistreri, G; Hellström, K; Golubtsov, AV; Jokitalo, E; Ahola, T (May 2011). "Assembly of alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells". Journal of Virology. 85 (10): 4739–51. doi:10.1128/JVI.00085-11. PMC 3126202. PMID 21389137.
- ^ Forrester NL, Palacios G, Tesh RB, Savji N, Guzman H, Sherman M, Weaver SC, Lipkin WI (December 2011). "Genome scale phylogeny of the Alphavirus genus suggests a marine origin". J Virol. 86 (5): 2729–38. doi:10.1128/JVI.05591-11. PMC 3302268. PMID 22190718.
- ^ Levinson RS, Strauss JH, Strauss EG (1990). "Complete sequence of the genomic RNA of O'nyong-nyong virus and its use in the construction of alphavirus phylogenetic trees". Virology. 175 (1): 110–123. doi:10.1016/0042-6822(90)90191-s. PMID 2155505.
- ^ "Virus Taxonomy: 2020 Release". International Committee on Taxonomy of Viruses (ICTV). March 2021. Retrieved 15 May 2021.
- ^ Tschá MK, Suzukawa AA, Gräf T, Piancini LD, da Silva AM, Faoro H, et al. (2019). "Identification of a novel alphavirus related to the encephalitis complexes circulating in southern Brazil". Emerging Microbes & Infections. 8 (1): 920–933. doi:10.1080/22221751.2019.1632152. PMC 6598490. PMID 31237479.
- ^ Powers AM, Brault AC, Shirako Y, Strauss EG, Kang W, Strauss JH, Weaver SC (November 2001). "Evolutionary relationships and systematics of the alphaviruses". J. Virol. 75 (21): 10118–31. doi:10.1128/JVI.75.21.10118-10131.2001. PMC 114586. PMID 11581380.
- ^ Lundström JO, Pfeffer M (November 2010). "Phylogeographic structure and evolutionary history of Sindbis virus". Vector Borne Zoonotic Dis. 10 (9): 889–907. doi:10.1089/vbz.2009.0069. PMID 20420530.
- ^ Weaver SC, Hagenbaugh A, Bellew LA, Netesov SV, Volchkov VE, Chang GJ, Clarke DK, Gousset L, Scott TW, Trent DW (November 1993). "A comparison of the nucleotide sequences of eastern and western equine encephalomyelitis viruses with those of other alphaviruses and related RNA viruses". Virology. 197 (1): 375–90. doi:10.1006/viro.1993.1599. PMID 8105605.
- ^ Weaver SC, Rico-Hesse R, Scott TW (1992). "Genetic diversity and slow rates of evolution in New World alphaviruses". Curr. Top. Microbiol. Immunol. Current Topics in Microbiology and Immunology. 176: 99–117. doi:10.1007/978-3-642-77011-1_7. ISBN 978-3-642-77013-5. PMID 1318187.
- ^ Comes, Jerome D.G.; Pijlman, Gorben P.; Hick, Tessy A.H. (14 June 2023). "Rise of the RNA machines – self-amplification in mRNA vaccine design". Trends in Biotechnology. 41 (11): 1417–1429. doi:10.1016/j.tibtech.2023.05.007. ISSN 0167-7799. PMC 10266560. PMID 37328401.
- ^ "Togaviridae". stanford.edu.
- ^ "ICTV Taxonomy List". Retrieved 5 May 2020.
- ^ "Aedes vigilax". NSW Arbovirus Surveillance & Vector Monitoring Program. The New South Wales Arbovirus Surveillance and Mosquito Monitoring Program. Archived from the original on 19 March 2018. Retrieved 5 June 2010.
Note that 'Ochlerotatus vigilax' prior to 2000, was known as 'Aedes vigilax'
- ^ Doherty RL, Carley JG, Best JC (May 1972). "Isolation of Ross River virus from man". The Medical Journal of Australia. 1 (21): 1083–4. doi:10.5694/j.1326-5377.1972.tb116646.x. PMID 5040017. S2CID 13048176.
- ^ Calisher CH (January 1994). "Medically important arboviruses of the United States and Canada". Clinical Microbiology Reviews. 7 (1): 89–116. doi:10.1128/CMR.7.1.89. PMC 358307. PMID 8118792.
- ^ Boughton CR, Hawkes RA, Naim HM (February 1988). "Illness caused by a Barmah Forest-like virus in New South Wales". The Medical Journal of Australia. 148 (3): 146–7. doi:10.5694/j.1326-5377.1988.tb112780.x. PMID 2828896. S2CID 30837260.
- ^ Tsetsarkin K, Higgs S, McGee CE, De Lamballerie X, Charrel RN, Vanlandingham DL (2006). "Infectious clones of Chikungunya virus (La Réunion isolate) for vector competence studies". Vector Borne and Zoonotic Diseases. 6 (4): 325–37. doi:10.1089/vbz.2006.6.325. PMID 17187566.
- ^ Lahariya C, Pradhan SK (December 2006). "Emergence of chikungunya virus in Indian subcontinent after 32 years: A review". Journal of Vector Borne Diseases. 43 (4): 151–60. PMID 17175699.